
A narrative review of BRAF alterations in human tumors: diagnostic and predictive implications
Introduction
B-Raf proto-oncogene, serine/threonine kinase (BRAF) represents one of the most commonly mutated and best characterized oncogenes in human tumorigenesis. The spectrum of tumors harboring BRAF mutations spans essentially every organ system and represents both indolent neoplasms and highly aggressive malignancies. Advances in clinical sequencing technologies have enabled routine detection of BRAF alterations beyond the V600E hotspot, leading to novel clinicopathologic correlations. Developments in targeted therapeutics against RAF and MEK pathway activation have led to a number of BRAF biomarker-driven clinical trials, beginning with targeted BRAF inhibitor monotherapy in melanoma, followed by exploratory pan-cancer basket trials, and ultimately culminating in specific targeted inhibitor combination therapies now approved for patients with advanced or metastatic BRAF V600E-mutated tumors. Understanding the spectrum and targetability of BRAF alterations is now fundamental to the practice of diagnostic and therapeutic oncology. This article will review the role of BRAF alterations in neoplasia, examine the recently defined classes of BRAF alteration with regard to downstream signaling and targetability, review select examples of BRAF targeting in clinical practice, and discuss molecular diagnostics for detection of BRAF mutations. We present the following article in accordance with the narrative review checklist (available at http://dx.doi.org/10.21037/pcm-2019-ppbt-02).
Methods
Literature used to inform the text herein was drawn from PubMed.gov from the National Library of Medicine and included full length manuscripts published in the English language between 1997 and 2020. Congress abstracts were identified through targeted searches of their sponsoring organization websites, where necessary. Regulatory documents were identified via Google search.
BRAF structure and function
BRAF belongs to the rapidly accelerated fibrosarcoma (RAF) family of serine/threonine kinases and functions as a Mitogen-activated pathway kinase kinase kinase (MAPKKK) in the MAP kinase/ERK signaling cascade. BRAF is normally triggered following ligand binding to receptor tyrosine kinases (RTKs) such as epidermal growth factor receptor (EGFR) or ERBB2. RTK phosphorylation leads to activation of the RAS-family of GTPases, which trigger dimerization of RAF family members and downstream activation of kinases including MEK1/2 and ERK1/2, leading to direct and indirect transcriptional regulation involved in cell survival and proliferation (1). Three distinct Raf genes have been described: ARAF, BRAF, and CRAF (RAF-1); all have been demonstrated to play essential roles in development and tumorigenesis. However, BRAF is maximally activated by oncogenic Ras signaling, whereas ARAF and CRAF appear to require Raf-dependent tyrosine phosphorylation (2). BRAF is located on chromosome 7q34; it is comprised of 18 exons and contains three regions that are conserved across the Raf family members: C1, containing the Raf-like Ras binding domain and an auto-inhibitor of the kinase domain (encoded by amino acids 150–290); C2, containing the serine and threonine-rich hinge region (encoded by amino acids 360–375); and C3, containing the protein tyrosine kinase domain (encoded by amino acids 457–717).
BRAF in disease
Germline BRAF mutations falling within the C1 domain and protein tyrosine kinase domain may give rise to Cardio-facio-cutaneous syndrome, which is associated with facial dysmorphism, mental retardation, and cardiac defects. Individuals with this disorder only rarely go on to develop malignancies (3,4). Somatic mutations in BRAF, on the other hand, are among the most common oncogenic alterations reported in humans and can be found in adult and pediatric cancer patients, in both solid and liquid tumors, and as apparent drivers of both highly aggressive and indolent neoplasms. Oncogenic mutations in BRAF are reported overall in 6% of human malignancies and are located principally within the C3 region (1). Although BRAF Val600Glu (V600E) mutations are the most well-recognized both for diagnostic and therapeutic purposes, over 200 oncogenic alterations have been reported in this gene with a range of implications for downstream pathway activation and targetability (5).
Oncogenesis versus senescence
BRAF mutations were first recognized as oncogenic in 2002, when investigators involved in The Cancer Genome Project described V600E mutations (annotated at the time as V599E) in a range of cancer types including in two-thirds of melanomas (6). However, these same mutations were also described in over 80% of benign nevi, suggesting that BRAF V600E is not sufficient to drive oncogenesis (7). In benign clonal processes such as nevi, BRAF oncogene-driven senescence blocks cellular proliferation via induction of the tumor suppressor p16INK4a and stable cell cycle arrest (7,8). The vast majority of benign nevi fail to progress to malignancy and some may even regress. In melanoma, in contrast, it appears that precursor cells first acquire hits in tumor suppressor genes such as CDKN2A and/or PTEN; in this context, subsequent mutations in MAPK genes, including BRAF, can drive malignant transformation (9). Indeed, it appears that in most tumor contexts with recurrent BRAF mutations, inactivation of a variety of tumor suppressor genes are required for transformation. Oncogene-induced senescence is also associated with telomere dysfunction; therefore in a subset of tumors the de-repression of hTERT gene expression, such as through TERT promoter mutation, restores telomerase activity and escape from senescence (10).
Besides melanocytic nevi, BRAF mutations are associated with a wide range of benign proliferations and neoplasms of low malignant potential impacting most organs of the body. These include endosalpingiosis (11), metanephric adenoma (12) and metanephric stromal tumors (12), papillary craniopharyngiomas (13), ganglioglioma (14), pituitary adenoma (15), bronchial adenomas/ciliated muconodular papillary tumor of the lung (16,17), Erdheim-Chester disease (18), Langerhans cell histiocytosis (19), and sessile serrated adenomas of the colon (20). Selected examples of indolent or low-grade neoplasms containing BRAF V600E mutations are described in more detail below.
Endosalpingiosis
Endosalpingiosis, defined as the presence of morphologically-benign glandular structures comprised of fallopian tube epithelium involving peritoneum or lymph nodes, is considered the precursor of ovarian low-grade serous neoplasms. Accordingly, similar driver oncogenic events are identified in both lesions, and in patients with both endosalpingiosis and low-grade serous neoplasms, a common KRAS G12/G13 or BRAF V600E mutation can be detected in both populations (11). In cell culture models of fallopian tube or ovarian surface epithelial cells, the presence of mutant KRAS or BRAF triggers growth arrest (11,21); consistent with this observation, the proliferation rate in endosalpingiosis is low. Additional defects in tumor suppressor pathways are likely necessary to drive the evolution from salpingiosis to clinically detectable serous tumors.
Histiocytoses
Erdheim chester disease (ECD) and Langerhans cell histiocytosis (LCH) are progressive, systemic neoplastic processes affecting both children and adults, currently characterized by the World Health Organization as inflammatory myeloid neoplasms. ECD is comprised of CD68+ histiocytes whereas LCH is comprised of CD1a+ CD207+ histiocytes; both are associated with multi-organ involvement. Fifteen percent of ECD patients also have LCH, a situation classified as “overlap histiocytosis” (22). Pulmonary LCH is uniquely associated with cigarette smoking, and the symptomatic and radiographic sequelae can often be managed through smoking cessation alone. The biologic relatedness of ECD and LCH is supported in part by evidence for similar genomic profiles, including frequent BRAF V600E mutations (in ~70% and ~60%, respectively, when using highly sensitive molecular techniques) and mutually exclusive MAP2K1 hotspot mutations (in 20% and 12%, respectively) (22,23). Other BRAF activating events including indels, duplication, or fusion are reported in a minor subset of LCH cases (24). BRAF V600E mutations were initially reported as sole alterations in histiocytosis (25); however, later studies have reported TP53-comutations (26) or loss of p16(INK4a) in aggressive cases (27), consistent with at least a two-hit model of tumorigenesis and the need for impaired tumor suppressor function to release the BRAF-mutated cells from senescence.
Colonic sessile serrated adenomas/polyps
BRAF V600E mutations are reported in approximately 60-80% of serrated neoplasms (28) but are absent in traditional adenomas of the colon. Sessile serrated adenomas/polyps (SSA/P) are thought to represent the benign precursor to CpG Island Promoter Methylation-high /microsatellite instability-high (MSI-H) colon cancers. Accordingly, SSA/P shows frequent BRAF V600E mutation, with acquisition of WNT pathway activity and widespread CpG island methylation, including MLH1 promoter methylation, during progression to cytologic dysplasia (29,30). A minority of BRAF-mutated SSP/A may progress down a microsatellite stable pathway via acquisition of tumor suppressor gene mutations including in TP53 and PTEN (31).
Overview of types of oncogenic alterations found in solid malignancies
Activating somatic BRAF alterations, including mutations, fusions, and amplifications, are reported in a diverse set of solid tumors. These are common in primary brain tumors, followed by non-follicular thyroid tumors, melanoma, and colorectal carcinoma. There is a long tail of other solid tumors that show mutations and/or fusions in 1–5% of cases (lung adenocarcinomas, acinar cell carcinomas of the pancreas, intrahepatic cholangiocarcinomas) or in fewer than 1% of cases (prostate and bladder carcinomas, high grade serous ovarian carcinoma) (32) (Figure 1).
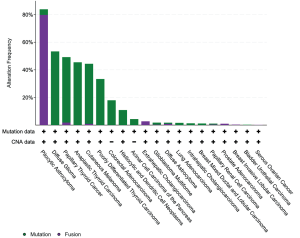
A subset of lower grade central nervous system tumors shows a particularly high frequency of BRAF activating alterations. BRAF V600E mutation is seen in nearly all papillary craniopharyngiomas. Pilocytic astrocytoma harbors a BRAF fusion or mutation in more than 80% of cases, and ganglioglioma and pleomorphic xanthoastrocytoma harbor BRAF fusions or mutations in up to 50% and 66% of cases, respectively. In contrast, BRAF mutations are rare in glioblastoma multiforme. BRAF alterations have variable and conflicting prognostic implications in lower grade primary brain tumors; however, identification of these changes can inform diagnosis and impact selection of BRAF targeted therapies (33). As a result, molecular analysis and/or fluorescence in situ hybridization maybe employed routinely in clinical practice to detect these changes and confirm a morphologic diagnosis.
Oncogenic activity of BRAF alterations
Mutations
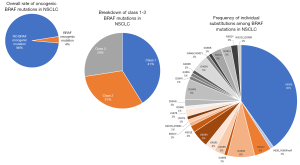
Oncogenic BRAF mutations are characterized based on whether they demonstrate kinase activity and require upstream RAS activity and BRAF dimerization. Kinase-activating mutations are independent of RAS signaling. Within this class of mutations, a subset signal as monomers (class 1) and others signal as constitutively active dimers (class 2). BRAF V600E falls into the class 1 activating group of mutants, along with other less common substitutions at this codon including V600K/D/R/M. Other small insertion deletion mutations involving the V600 codon are rare, but at least isolated examples may stabilize the activated kinase in a similar fashion to other class 1 mutants (33). Class 1 mutations tend to occur in a mutually exclusive fashion with other oncogenic driver alterations (e.g., KRAS, EGFR, ALK fusions, etc.), at least in the pre-treatment setting (34). Class 2 mutants cluster at codons 601, 597, 469 and 464 (5). Finally, a subset of BRAF mutations demonstrates low to absent kinase (kinase-dead) activity, and activation of downstream signaling is dependent on RAS activity. These so-called class 3 mutants are scattered throughout hotspots in exons 11 and 15 and represent up to 30% of BRAF mutations observed in colorectal carcinoma and up to 30% of BRAF mutations observed in non-small cell lung carcinoma (1) (Figure 2), occasionally in tandem with oncogenic mutations in RAS family member genes. Mechanistically, these RAS-activated kinase-low/dead mutants appear to heterodimerize with CRAF to trigger ERK signaling and thereby amplify other inputs into the ERK pathway (5). While class 1 (V600) mutants are sensitive to targeted RAF inhibitors, class 2 mutants may require dual inhibition of RAF and downstream MEK signaling (36), and class 3 mutants appear responsive to MEK inhibitors (5).
Fusions and other structural variants
Oncogenic BRAF fusions were first reported in papillary thyroid tumors, enriched in individuals exposed to radiation following the Chernobyl nuclear accident (37). AKAP9-BRAF was the first characterized fusion, comprised of the first 8 exons of AKAP9 and the C-terminal portion of BRAF including exons 9–18 and resulting from paracentric chromosomal inversion event on the long arm of chromosome 7. This event led to loss of the CR1 and CR2 regulatory domains within the N-terminal portion of BRAF, with retention of the kinase domain. Consistent with loss of the regulatory region of the protein, the fusion product demonstrates constitutive RAF kinase activation and transforming ability. Structurally similar fusions were subsequently described in pilocytic astrocytomas, where a duplication event on 7q34 leads to fusion of the N terminus of KIAA1549 and the C terminus of BRAF beginning at either exons 9 or 11 (38). Other less common but recurrent fusions include FAM131B-BRAF, resulting from an interstitial deletion on 7q (39) in pilocytic astrocytoma and SND1-BRAF mutations in lung adenocarcinoma and pancreatic acinar cell carcinomas (40). Over 50 additional fusion partners have been identified in one to two reports each in a spectrum of tumors including pilocytic astrocytoma, spitzoid melanomas, and other solid tumor types at an exceptionally low frequency (41,42). Internal tandem duplications of the BRAF kinase domain and/or intragenic deletion of the N-terminal regulatory domains have been reported rarely in melanoma and infantile fibrosarcoma (43,44).
Fusion events are considered Class 2 alterations; the loss of the N-terminal regulatory domains enables RAS-independent homodimerization, which is required for kinase activation in this class of mutants. As with the point mutations in this class, BRAF fusions drive MAPK pathway signaling but are resistant to first generation BRAF inhibitors such as vemurafenib (45). Next generation RAF inhibitors that inhibit BRAF homo- and heterodimers may have more activity in BRAF-fusion driven tumors (46).
Therapeutic implications of BRAF mutations
Melanoma
Oncogenic mutations in BRAF are reported in over 40% of cutaneous melanomas, are enriched in tumors arising on skin without chronic sun damage and are clustered in and around codon V600 (47). The most common substitution is V600E followed by V600K; other substitutions including V600R/M/D/G and small indels at this position are uncommon (48). Clinical trials of single agent vemurafenib in patients with metastatic melanoma and BRAF V600E/K mutations showed objective response rates of around 50% (49-51). Overall survival with vemurafenib therapy was improved relative to standard of care at the time of the published monotherapy trials (52) but was limited by development of resistance (53,54). Melanomas exposed to BRAF inhibitors employ heterogeneous mechanisms to reactivate MAPK and alternative pathways (55). Combination RAF and MEK therapy (dabrafenib, trametinib) provides improved overall survival relative to vemurafenib monotherapy and >60% response rate (56), but shows only modest efficacy in patients with BRAF inhibitor-refractory melanoma following monotherapy (57). Current (2020) clinical testing and treatment guidelines recommend BRAF mutational testing for any patient with stage III or IV cutaneous melanoma; detection of a BRAF V600-activating mutation justifies use of BRAF-MEK therapies including dabrafenib/trametinib, vemurafenib/cobimetinib, or encorafenib/binimetinib. These therapies may be used in the first line or after progression on immunotherapy with PD-1 inhibitors (58). Mutations occurring at codons L597 and K601 may also predict response to MEK or combination MEK/RAF inhibitors, however other exon 11 and 15 mutations do not (36). Post-hoc analyses of clinical trials of the PD-1 inhibitor pembrolizumab demonstrate efficacy of this immunotherapeutic irrespective of BRAF V600 mutational status or prior treatment with BRAF/MEK inhibitors (59).
Targeting BRAF beyond melanoma
Following positive trials of BRAF inhibitor therapy in melanomas, basket trials were opened to enroll patients across various BRAF V600-mutated nonmelanoma cancers to determine if targeted BRAF inhibitors might be a promising approach across diagnoses. Initial results for vemurafenib monotherapy were disappointing, however, with most activity seen in NSCLC and histiocytosis. Occasional durable responses were seen in other tumor types including cholangiocarcinoma, anaplastic thyroid carcinoma, and ovarian cancer, and no activity was observed for combined vemurafenib and cetuximab in colon cancer (60). Given these at best modest initial results, investigators began instead looking to combinatorial therapy in a variety of clinical contexts.
Colon
BRAF V600E mutations are reported in 10% of colorectal carcinomas, are enriched in right sided tumors and those with sporadic mismatch repair deficiency due to MLH1 promoter methylation, and serve as exclusionary criteria for selection of patients for treatment with anti-EGFR (such as cetuximab). This is based on the assumption that activation of RAF/RAS kinase members such as KRAS, NRAS and BRAF leads to constitutive MAPK pathway activity independent of RTK signaling, such as through EGFR (1). In retrospective studies of colon cancer patients treated with anti-EGFR monoclonal antibodies, BRAF V600 mutations serve as negative predictors of response (61,62). Preclinical studies have demonstrated that inhibition of BRAF V600E in colon cancer leads to feedback activation of EGFR and bypass activation of MAPK pathway via CRAF. Indeed, the combination of anti-BRAF and anti-EGFR agents have a synergistic effect in killing of BRAF mutant colon cancer cells (63,64). Phase I/II studies of combined MEK and BRAF inhibitors showed a modest improvement in response over BRAF inhibitors alone in patients with BRAF V600E mutant colon cancers (65). Trials of triplet BRAF, EGFR and MEK inhibition in patients with BRAF V600E mutant metastatic colorectal carcinoma (BEACON) demonstrated tolerability and improved efficacy over standard of care therapy (66,67). In 2018, the FDA granted breakthrough therapy designation to BRAF, MEK, and EGFR inhibitor combination (encorafenib, binimetinib, and cetuximab, respectively) as second line therapy (68). Patients in the BEACON study receiving doublet encorafenib and cetuximab therapy in the second or third lines also showed significant improvements in response rates and survival relative to standard of care, leading to FDA approval in 2020 (69). Non-V600 mutations are not so clearly associated with lack of response to anti-EGFR therapies (70,71). In theory, a subset of tumors with class 3 mutations lacking co-mutations in other RAS pathway members and retaining dependence on RTK signaling may derive benefit from anti-EGFR therapy (72).
Sixty percent of sporadic MLH1 promoter methylated mismatch repair-deficient (MMR-D)/MSI-H colorectal carcinomas have a BRAF V600E mutation; this contrasts with only about 1% of colon cancers arising in patients with germline MMR mutations (73). Therefore, knowledge of the BRAF V600E status can inform the likelihood of Lynch Syndrome-associated versus sporadic colorectal carcinoma. BRAF mutation, taken together with MMR/MSI status, is a recognized prognostic indicator in colorectal carcinoma. MSI-H/BRAF wild type tumors have the best survival outcomes, whereas microsatellite stable (MSS)/BRAF mutant tumors are associated with poor prognosis. MSS/BRAF wild type tumors appear to have intermediate outcomes (74,75). Besides its association with BRAF mutation in colon cancer, MMR-D status predicts responsiveness to PD-1 inhibitor therapy (76,77). In a phase 3 trial of the PD-1 inhibitor pembrolizumab versus standard of care frontline chemotherapy for patients with MMR-D/MSI-H colorectal carcinoma, pembrolizumab more than doubled progression free survival with a reduction in severe adverse effects as compared to cytotoxic therapy (78). This led to an FDA approval for pembrolizumab as first line treatment of colorectal carcinoma with MMR-D/MSI-H status, irrespective of BRAF or other oncogenic driver mutation status (79).
Lung
BRAF exhibits diverse mutations in NSCLC; just under half of reported oncogenic BRAF mutations occur at V600E; the remainder are distributed predominantly within exons 11 and 15 and are associated with class 2 or 3 activity (Figure 2). The FDA has approved combination dabrafenib and trametinib (RAF and MEK inhibitors) in the first line of therapy for patients with advanced/metastatic BRAF V600E-mutated NSCLC (80). This combination is associated with improved response, progression free survival, and overall survival as compared to chemotherapy (81), and a subset of patients experiences durable benefit, with over a third of patients treated in the first line setting surviving more than three years (82). Durable response to this combination therapy may be related to the simplicity of the NSCLC genome, with more complex genetic changes and co-mutations predicting shorter duration of benefit (82). The clinical characteristics of patients with BRAF V600E mutations are heterogeneous, including never smokers and former/current smokers; adenocarcinoma is the most common histology observed. In contrast to EGFR/ALK/ROS1-altered lung carcinomas, which tend to show only very limited response to immunotherapies, tumors with BRAF mutations, including both V600E and non-V600E mutations, do show response to immune checkpoint inhibitors (83). Combination RAF/MEK inhibitor therapy is not approved for non-V600E BRAF mutations in light of preclinical data suggesting limited efficacy of approved BRAF inhibitors in tumors with mutations leading to BRAF dimerization. In NSCLC, class 2 and 3 BRAF mutations are more likely to occur in tandem with mutations in KRAS and appear to be associated with more aggressive disease (35). Trials of MEK and pan-RAF inhibitors have been attempted in this context but have been limited by drug toxicity (84).
Other rare tumor types
Combined BRAF and MEK inhibitor therapies have been approved for use in BRAF V600E mutated anaplastic thyroid carcinoma (85), which represents about 1–2% of thyroid cancers overall and harbors this mutation in nearly 50% of cases. Over two-thirds of BRAF-mutated anaplastic thyroid carcinoma patients treated with this regimen respond, with a 12-month overall survival of 80%, dramatically greater than historic 1 year survival rates of 20–40% (86).
The BRAF inhibitor vemurafenib was approved for BRAF V600-mutated ECD in 2017, based on results of the VE Basket trial that enrolled 22 adult ECD patients; over half of patients responded and reported an improvement in neurologic symptoms and pain (87,88). Responses to BRAF inhibition with or without MEK inhibition have been reported in other rare tumor types including BRAF V600E-mutated ameloblastoma, malignant glomus tumors, cholangiocarcinoma, salivary gland adenocarcinoma, and glioblastoma (89-91).
Diagnostic tools for BRAF mutation detection
A wide variety of methodologies have been employed for detection of BRAF V600 mutations, and it is important to recognize the strengths and deficiencies of different strategies. Beginning with the approval of vemurafenib therapy for BRAF V600-mutated melanoma in 2011, a trend of companion diagnostic approvals began to dictate formally accepted practices for clinical mutation detection. The Roche cobas 4800 BRAF V600 Mutation test was approved as an in vitro diagnostic for vemurafenib as monotherapy as well as in combination with cobimetinib (92). This targeted PCR-based assay quickly came under criticism because of its inability to detect less common dinucleotide substitutions leading to V600E/K/R mutations and its suboptimal sensitivity relative to unbiased sequencing based methods (93). At the same time, the highly standardized cobas test appeared to generate fewer invalid results relative to Sanger sequencing and primer-extension/fragment analysis assays (94) and showed superior sensitivity to Sanger sequencing for samples with mutant allele fraction of under 25% (95). A practical downside to companion diagnostic usage in clinical laboratories is the capital expenditure required to acquire platform-specific instrumentation, creating a barrier to implementation in small laboratories with tighter budgets. Many molecular laboratories already have instruments suited to analysis of the specific clinically relevant mutation and little incentive to purchase another piece of potentially duplicative equipment. Further complicating this space, the FDA more recently approved a different BRAF assay—the therascreen BRAF V600E Rotor-Gene Q PCR kit, requiring dedicated instrumentation—for selection of colorectal carcinoma patients for BRAF and EGFR inhibitor therapy (96). Many molecular diagnosticians will ask why two different FDA-approved platforms should be required to generate information about the same mutational change.
Analyses of over 1000 national proficiency test results between 2011–2015 indicated that acceptable (correct) responses were generated for 96.6% of BRAF results in laboratories relying on laboratory developed tests as compared to 93% for FDA-approved companion diagnostics including the Roche cobas BRAF and bioMerieux THxID BRAF tests; the lower performance was driven by the Roche assay and attributed to its relative insensitivity for detection of the c.1798_1799delGTinsAA (p.V600K) dinucleotide substitution (97). In another proficiency testing analysis examining use of next generation sequencing versus non-NGS practices for mutation detection from 2014–2017, laboratories that employed NGS methodology showed a significantly greater likelihood of achieving a correct result for BRAF testing, driven again by the ability to more reliably detect the V600K change (98). Ultimately, evidence for greater cost-effectiveness and shorter overall turnaround time for NGS assays (99) that can deliver information for many more potential targets across tumor types, as well as availability of a number of FDA-approved NGS assays (100), will likely continue to push the field away from reliance on single gene assays.
Immunohistochemistry (IHC)
Mutation-specific BRAF V600E IHC using the VE1 clone is an accepted screening tool for melanoma. Relative to molecular testing, VE1 IHC ranges 85–97% sensitivity and 96-100% specificity, depending on the defined comparator (101,102). The short turnaround time afforded by IHC testing can enable patients to start BRAF inhibitor therapy earlier than may be possible when relying on molecular testing—this can offer clinical benefit to patients with rapidly progressive disease. However, given the less-than-perfect performance characteristics, confirmatory molecular testing is generally recommended (58). Metastatic lesions and pigmented tumors most commonly give rise to IHC—molecular discrepancies (103). Decalcification of metastatic melanoma in bone samples can lead to falsely negative IHC and/or molecular testing results and may be a source of discrepancy between these methods (104,105). VE1 IHC is also insensitive to the BRAF V600K (and other non-V600E mutations); therefore, mutational profiling is required to detect these alterations (103).
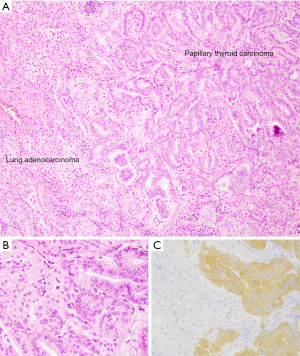
For thyroid carcinoma, VE1 IHC has been validated for detection of BRAF V600E mutations in tissue and cytology cell block specimens (103,106); however, the performance of the antibody in direct smears and liquid-based preparations, particularly those with obscuring elements, is limited (107,108). The specificity of the VE1 IHC staining is illustrated in a collision tumor including a BRAF-wild type lung adenocarcinoma and a metastatic BRAF V600E-mutated papillary thyroid carcinoma (Figure 3).
BRAF IHC has been proposed as a tool for detection of serrated lesions of the colon (109) as well as to screen colorectal cancers for BRAF V600E mutations (110). Early studies of this IHC antibody in cohorts of colon neoplasms gave conflicting results, with very promising results from a laboratory using hybridoma supernatants (109), but evidence of poor sensitivity and specificity for BRAF-mutated colon cancers from a variety of groups using commercially available antibodies (111-113). Other groups observed that VE1 IHC and molecular testing could show high concordance but required rigorous optimization of the IHC assay and even so resulted in a substantial number of cases with weak staining requiring confirmation via molecular methods (103,114). Access to molecular assays, including comprehensive next generation sequencing tests that deliver information on a wide variety of potential biomarkers simultaneously, may supersede BRAF IHC, given that it delivers information on only one therapeutically relevant variable. In theory, however, BRAF VE1 IHC combined with MMR protein evaluation can provide most of the necessary information for first or later line treatment decisions in patients with colon adenocarcinoma (see Therapeutic Implications, above).
Detection of BRAF V600E mutation in non-small cell lung carcinoma, particularly lung adenocarcinoma, is now an indication for first line therapy with combined BRAF/MEK inhibitors. This clinical indication has driven a renewed interest in use of VE1 IHC for rapid screening/detection of BRAF V600E mutations in this tumor type. Mutations occurring at the V600 codon other than V600E are exceptionally rare in NSCLC; therefore, the clinical false negativity issues that impact melanoma testing are not relevant in this tumor type. Given the rarity of BRAF V600E mutation in NSCLC, earlier studies had only small numbers of true positive cases (115,116), so despite relatively robust performance characteristics, the data was limited and this testing approach was generally discouraged by clinical testing guidelines (117). A number of more recent studies using larger molecularly defined cohorts have confirmed the utility of VE1 testing in NSCLC, with sensitivities ranging from 96.6–100% and specificities ranging from 98.6–100%. Thus, it may be expected that VE1 IHC can be implemented into routine IHC-based predictive biomarker screening (118,119).
Summary
To summarize, BRAF is a central member of the MAPK signaling pathway, activation of which drives pro-survival and proliferation programs. Critically, activation of BRAF alone appears insufficient to drive malignant tumor behavior, rather, it enables a program of oncogene-driven senescence that manifests as indolent clonal processes such as benign nevi or adenomas arising at various body locations. It appears that a second hit is required for malignant transformation in the setting of a BRAF activating event; genes implicated in that transformation step include CDKN2A, PTEN, and TP53. Based on available functional evidence, BRAF genomic alterations, which include the well-known point mutations (including at V600), as well as small indels and larger structural variants, are considered Class 1, 2, or 3, based on their ability to trigger downstream signaling as monomers, dimers, or through RAS dependence. This classification has important implications for the efficacy of BRAF targeted therapies, which appear most potent against those tumors harboring Class 1 mutations.
BRAF is mutated or otherwise altered in tumors arising in essentially every body site in a variety of genomic backgrounds, giving rise to a tremendous diversity of pathologic and clinical neoplastic manifestation. For patients with advanced disease, the efficacy of RAF-targeted therapy is dictated by the context in which the BRAF mutation is found, with greatest clinical efficacy coming from combination therapies that target both RAF and other upstream (EGFR) or downstream (MEK/ERK) signals. Given the broad clinical implications for BRAF alterations in human neoplasia, accurate and sensitive biomarker testing is essential in practice. An array of testing approaches has been developed and optimized, ranging from targeted PCR-based molecular diagnostics to BRAF V600E mutation-specific IHC to high throughput next generation sequencing. These varied methods all have their strengths and weaknesses, but molecular methods tend to show very similar performance characteristics, irrespective of their status as laboratory tests or FDA-labeled companion diagnostics. Laboratories may choose to employ multiple testing approaches to leverage respective strengths, e.g., turnaround time, sensitivity, or breadth.
Ongoing research into therapies for patients with BRAF-driven malignancies will likely continue to focus on optimizing the combination of targeted inhibitors and will leverage immunotherapy, at least in a subset of BRAF-driven tumors. However, our understanding of the intrinsic and acquired resistance to these therapies is still immature, and in all likelihood analyses of tumors that moves beyond the genome, and into the methylome, proteome, and beyond, will be required to more fully dissect the pathways responsible.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors Mari Mino-Kenudson and Yin (Rex) P. Hung for the series “Predictive and Prognostic Biomarkers in Tumors” published in Precision Cancer Medicine. The article has undergone external peer review.
Reporting Checklist: The author has completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/pcm-20-39
Conflicts of Interest: The author has completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm-20-39). The series “Predictive and Prognostic Biomarkers in Tumors” was commissioned by the editorial office without any funding or sponsorship. LMS reports grants from Genentech, personal fees from EMD Serono, personal fees from Foghorn Therapeutics, and personal fees from AstraZeneca, outside the submitted work. The author has no other conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects for the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018;37:3183-99. [Crossref] [PubMed]
- Marais R, Light Y, Paterson HF, et al. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem 1997;272:4378-83. [Crossref] [PubMed]
- Niemeyer CM. RAS diseases in children. Haematologica 2014;99:1653-62. [Crossref] [PubMed]
- Kratz CP, Franke L, Peters H, et al. Cancer spectrum and frequency among children with Noonan, Costello, and cardio-facio-cutaneous syndromes. Br J Cancer 2015;112:1392-7. [Crossref] [PubMed]
- Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234-8. [Crossref] [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [Crossref] [PubMed]
- Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19-20. [Crossref] [PubMed]
- Michaloglou C, Vredeveld LC, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature 2005;436:720-4. [Crossref] [PubMed]
- Damsky WE, Bosenberg M. Melanocytic nevi and melanoma: unraveling a complex relationship. Oncogene 2017;36:5771-92. [Crossref] [PubMed]
- Patel PL, Suram A, Mirani N, et al. Derepression of hTERT gene expression promotes escape from oncogene-induced cellular senescence. Proc Natl Acad Sci U S A 2016;113:E5024-33. [Crossref] [PubMed]
- Chui MH, Shih IM. Oncogenic BRAF and KRAS mutations in endosalpingiosis. J Pathol 2020;250:148-58. [Crossref] [PubMed]
- Chan E, Stohr BA, Croom NA, et al. Molecular characterisation of metanephric adenomas beyond BRAF: genetic evidence for potential malignant evolution. Histopathology 2020;76:1084-90. [Crossref] [PubMed]
- Martinez-Barbera JP, Andoniadou CL. Biological Behaviour of Craniopharyngiomas. Neuroendocrinology 2020;110:797-804. [Crossref] [PubMed]
- Koelsche C, Wohrer A, Jeibmann A, et al. Mutant BRAF V600E protein in ganglioglioma is predominantly expressed by neuronal tumor cells. Acta Neuropathol 2013;125:891-900. [Crossref] [PubMed]
- Chen J, Jian X, Deng S, et al. Identification of recurrent USP48 and BRAF mutations in Cushing's disease. Nat Commun 2018;9:3171. [Crossref] [PubMed]
- Chang JC, Montecalvo J, Borsu L, et al. Bronchiolar Adenoma: Expansion of the Concept of Ciliated Muconodular Papillary Tumors With Proposal for Revised Terminology Based on Morphologic, Immunophenotypic, and Genomic Analysis of 25 Cases. Am J Surg Pathol 2018;42:1010-26. [Crossref] [PubMed]
- Kim L, Kim YS, Lee JS, et al. Ciliated muconodular papillary tumor of the lung harboring BRAF V600E mutation and p16(INK4a) overexpression without proliferative activity may represent an example of oncogene-induced senescence. J Thorac Dis 2017;9:E1039-44. [Crossref] [PubMed]
- Cavalli G, Biavasco R, Borgiani B, et al. Oncogene-induced senescence as a new mechanism of disease: the paradigm of erdheim-chester disease. Front Immunol 2014;5:281. [Crossref] [PubMed]
- Roden AC, Hu X, Kip S, et al. BRAF V600E expression in Langerhans cell histiocytosis: clinical and immunohistochemical study on 25 pulmonary and 54 extrapulmonary cases. Am J Surg Pathol 2014;38:548-51. [Crossref] [PubMed]
- Kriegl L, Neumann J, Vieth M, et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol 2011;24:1015-22. [Crossref] [PubMed]
- Zeppernick F, Ardighieri L, Hannibal CG, et al. BRAF mutation is associated with a specific cell type with features suggestive of senescence in ovarian serous borderline (atypical proliferative) tumors. Am J Surg Pathol 2014;38:1603-11. [Crossref] [PubMed]
- Haroche J, Cohen-Aubart F, Amoura Z. Erdheim-Chester disease. Blood 2020;135:1311-8. [Crossref] [PubMed]
- Diamond EL, Durham BH, Haroche J, et al. Diverse and Targetable Kinase Alterations Drive Histiocytic Neoplasms. Cancer Discov 2016;6:154-65. [Crossref] [PubMed]
- Rodriguez-Galindo C, Allen CE. Langerhans cell histiocytosis. Blood 2020;135:1319-31. [Crossref] [PubMed]
- Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood 2014;124:3007-15. [Crossref] [PubMed]
- McGinnis LM, Nybakken G, Ma L, et al. Frequency of MAP2K1, TP53, and U2AF1 Mutations in BRAF-mutated Langerhans Cell Histiocytosis: Further Characterizing the Genomic Landscape of LCH. Am J Surg Pathol 2018;42:885-90. [Crossref] [PubMed]
- Chilosi M, Facchetti F, Calio A, et al. Oncogene-induced senescence distinguishes indolent from aggressive forms of pulmonary and non-pulmonary Langerhans cell histiocytosis. Leuk Lymphoma 2014;55:2620-6. [Crossref] [PubMed]
- O'Brien MJ, Yang S, Mack C, et al. Comparison of microsatellite instability, CpG island methylation phenotype, BRAF and KRAS status in serrated polyps and traditional adenomas indicates separate pathways to distinct colorectal carcinoma end points. Am J Surg Pathol 2006;30:1491-501. [Crossref] [PubMed]
- Murakami T, Mitomi H, Saito T, et al. Distinct WNT/beta-catenin signaling activation in the serrated neoplasia pathway and the adenoma-carcinoma sequence of the colorectum. Mod Pathol 2015;28:146-58. [Crossref] [PubMed]
- Yozu M, Kem M, Cenaj O, et al. Loss of expression of MLH1 in non-dysplastic crypts is a harbinger of neoplastic progression in sessile serrated adenomas/polyps. Histopathology 2019;75:376-84. [Crossref] [PubMed]
- Murakami T, Akazawa Y, Yatagai N, et al. Molecular characterization of sessile serrated adenoma/polyps with dysplasia/carcinoma based on immunohistochemistry, next-generation sequencing, and microsatellite instability testing: a case series study. Diagn Pathol 2018;13:88. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Maraka S, Janku F. BRAF alterations in primary brain tumors. Discov Med 2018;26:51-60. [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Dagogo-Jack I, Martinez P, Yeap BY, et al. Impact of BRAF Mutation Class on Disease Characteristics and Clinical Outcomes in BRAF-mutant Lung Cancer. Clin Cancer Res 2019;25:158-65. [Crossref] [PubMed]
- Dankner M, Lajoie M, Moldoveanu D, et al. Dual MAPK Inhibition Is an Effective Therapeutic Strategy for a Subset of Class II BRAF Mutant Melanomas. Clin Cancer Res 2018;24:6483-94. [Crossref] [PubMed]
- Ciampi R, Knauf JA, Kerler R, et al. Oncogenic AKAP9-BRAF fusion is a novel mechanism of MAPK pathway activation in thyroid cancer. J Clin Invest 2005;115:94-101. [Crossref] [PubMed]
- Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 2008;68:8673-7. [Crossref] [PubMed]
- Cin H, Meyer C, Herr R, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol 2011;121:763-74. [Crossref] [PubMed]
- Chmielecki J, Hutchinson KE, Frampton GM, et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov 2014;4:1398-405. [Crossref] [PubMed]
- Ross JS, Wang K, Chmielecki J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer 2016;138:881-90. [Crossref] [PubMed]
- Archer Quiver Fusion Database [updated May 1, 2018]. Available online: http://quiver.archerdx.com/results?query=braf.
- Kemper K, Krijgsman O, Kong X, et al. BRAF(V600E) Kinase Domain Duplication Identified in Therapy-Refractory Melanoma Patient-Derived Xenografts. Cell Rep 2016;16:263-77. [Crossref] [PubMed]
- Wegert J, Vokuhl C, Collord G, et al. Recurrent intragenic rearrangements of EGFR and BRAF in soft tissue tumors of infants. Nat Commun 2018;9:2378. [Crossref] [PubMed]
- Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A 2013;110:5957-62. [Crossref] [PubMed]
- Yao Z, Gao Y, Su W, et al. RAF inhibitor PLX8394 selectively disrupts BRAF dimers and RAS-independent BRAF-mutant-driven signaling. Nat Med 2019;25:284-91. [Crossref] [PubMed]
- Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135-47. [Crossref] [PubMed]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015;161:1681-96. [Crossref] [PubMed]
- Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809-19. [Crossref] [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [Crossref] [PubMed]
- Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012;366:707-14. [Crossref] [PubMed]
- McArthur GA, Chapman PB, Robert C, et al. Safety and efficacy of vemurafenib in BRAF(V600E) and BRAF(V600K) mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol 2014;15:323-32. [Crossref] [PubMed]
- Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973-7. [Crossref] [PubMed]
- Sun C, Wang L, Huang S, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014;508:118-22. [Crossref] [PubMed]
- Shi H, Hugo W, Kong X, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov 2014;4:80-93. [Crossref] [PubMed]
- Robert C, Karaszewska B, Schachter J, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015;372:30-9. [Crossref] [PubMed]
- Johnson DB, Flaherty KT, Weber JS, et al. Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol 2014;32:3697-704. [Crossref] [PubMed]
- Cutaneous Melanoma 2020 [updated May 18, 2020; cited 2020]. Version 3.2020. Available online: https://www.nccn.org/professionals/physician_gls/pdf/cutaneous_melanoma.pdf
- Puzanov I, Ribas A, Robert C, et al. Association of BRAF V600E/K Mutation Status and Prior BRAF/MEK Inhibition With Pembrolizumab Outcomes in Advanced Melanoma: Pooled Analysis of 3 Clinical Trials. JAMA Oncol 2020;6:1256-64. [Crossref] [PubMed]
- Hyman DM, Puzanov I, Subbiah V, et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N Engl J Med 2015;373:726-36. [Crossref] [PubMed]
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 2010;11:753-62. [Crossref] [PubMed]
- Shinozaki E, Yoshino T, Yamazaki K, et al. Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study. Br J Cancer 2017;117:1450-8. [Crossref] [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [Crossref] [PubMed]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227-35. [Crossref] [PubMed]
- Corcoran RB, Atreya CE, Falchook GS, et al. Combined BRAF and MEK Inhibition With Dabrafenib and Trametinib in BRAF V600-Mutant Colorectal Cancer. J Clin Oncol 2015;33:4023-31. [Crossref] [PubMed]
- Corcoran RB, Andre T, Atreya CE, et al. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAF(V600E)-Mutant Colorectal Cancer. Cancer Discov 2018;8:428-43. [Crossref] [PubMed]
- Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med 2019;381:1632-43. [Crossref] [PubMed]
- Huijberts SC, van Geel RM, Bernards R, et al. Encorafenib, binimetinib and cetuximab combined therapy for patients with BRAFV600E mutant metastatic colorectal cancer. Future Oncol 2020;16:161-73. [Crossref] [PubMed]
- FDA approves encorafenib in combination with cetuximab for metastatic colorectal cancer with a BRAF V600E mutation 2020 [updated 4/9/2020]. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-encorafenib-combination-cetuximab-metastatic-colorectal-cancer-braf-v600e-mutation
- Shinozaki E, Yoshino T, Tsuchihara K. Reply to `Comment on `Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study''. Br J Cancer 2018;118:1278-9. [Crossref] [PubMed]
- Jones JC, Renfro LA, Al-Shamsi HO, et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol 2017;35:2624-30. [Crossref] [PubMed]
- Dankner M, Rose AAN. Comment on 'Clinical significance of BRAF non-V600E mutations on the therapeutic effects of anti-EGFR monoclonal antibody treatment in patients with pretreated metastatic colorectal cancer: the Biomarker Research for anti-EGFR monoclonal Antibodies by Comprehensive Cancer genomics (BREAC) study'. Br J Cancer 2018;118:1276-7. [Crossref] [PubMed]
- Parsons MT, Buchanan DD, Thompson B, et al. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet 2012;49:151-7. [Crossref] [PubMed]
- Lochhead P, Kuchiba A, Imamura Y, et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst 2013;105:1151-6. [Crossref] [PubMed]
- Phipps AI, Alwers E, Harrison T, et al. Association Between Molecular Subtypes of Colorectal Tumors and Patient Survival, Based on Pooled Analysis of 7 International Studies. Gastroenterology 2020;158:2158-68.e4. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Le DT, Kim TW, Van Cutsem E, et al. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J Clin Oncol 2020;38:11-9. [Crossref] [PubMed]
- Andre T, Shiu KK, Kim TW, et al. Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: The phase 3 KEYNOTE-177 Study. J Clin Oncol 2020;38:LBA4. [Crossref]
- FDA approves pembrolizumab for first-line treatment of MSI-H/dMMR colorectal cancer 2020 [updated 6/30/2020]. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases/fda-approves-pembrolizumab-first-line-treatment-msi-hdmmr-colorectal-cancer
- Odogwu L, Mathieu L, Blumenthal G, et al. FDA Approval Summary: Dabrafenib and Trametinib for the Treatment of Metastatic Non-Small Cell Lung Cancers Harboring BRAF V600E Mutations. Oncologist 2018;23:740-5. [Crossref] [PubMed]
- Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307-16. [Crossref] [PubMed]
- Planchard D, Besse B, Groen H, et al. Updated overall survival (OS) and genomic analysis from a single-arm phase II study of dabrafenib (D) + trametinib (T) in patients (pts) with BRAF V600E mutant (Mut) metastatic non-small cell lung cancer (NSCLC). J Clin Oncol 2020;38:9593. [Crossref]
- Dudnik E, Peled N, Nechushtan H, et al. BRAF Mutant Lung Cancer: Programmed Death Ligand 1 Expression, Tumor Mutational Burden, Microsatellite Instability Status, and Response to Immune Check-Point Inhibitors. J Thorac Oncol 2018;13:1128-37. [Crossref] [PubMed]
- Dagogo-Jack I, Awad MM, Shaw AT. BRAF Mutation Class and Clinical Outcomes-Response. Clin Cancer Res 2019;25:3189. [Crossref] [PubMed]
- FDA approves new uses for two drugs administered together for the treatment of BRAF-positive anaplastic thyroid cancer 2018 [updated 5/4/2018]. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-new-uses-two-drugs-administered-together-treatment-braf-positive-anaplastic-thyroid
- Subbiah V, Kreitman RJ, Wainberg ZA, et al. Dabrafenib and Trametinib Treatment in Patients With Locally Advanced or Metastatic BRAF V600-Mutant Anaplastic Thyroid Cancer. J Clin Oncol 2018;36:7-13. [Crossref] [PubMed]
- Diamond EL, Subbiah V, Lockhart AC, et al. Vemurafenib for BRAF V600-Mutant Erdheim-Chester Disease and Langerhans Cell Histiocytosis: Analysis of Data From the Histology-Independent, Phase 2, Open-label VE-BASKET Study. JAMA Oncol 2018;4:384-8. [Crossref] [PubMed]
- Oneal PA, Kwitkowski V, Luo L, et al. FDA Approval Summary: Vemurafenib for the Treatment of Patients with Erdheim-Chester Disease with the BRAFV600 Mutation. Oncologist 2018;23:1520-4. [Crossref] [PubMed]
- Brunet M, Khalifa E, Italiano A. Enabling Precision Medicine for Rare Head and Neck Tumors: The Example of BRAF/MEK Targeting in Patients With Metastatic Ameloblastoma. Front Oncol 2019;9:1204. [Crossref] [PubMed]
- Cuviello A, Goyal A, Zick A, et al. Sporadic Malignant Glomus Tumor of the Brachial Plexus With Response to Targeted Therapy Directed Against Oncogenic BRAF. JCO Precis Oncol. 2018;2018.
- Groisberg R, Hong DS, Roszik J, et al. Clinical Next-Generation Sequencing for Precision Oncology in Rare Cancers. Mol Cancer Ther 2018;17:1595-601. [Crossref] [PubMed]
- Roche Cobas DNA Sample Preparation Kit, COBAS 4800 BRAF V600 mutation test 2020 [updated 7/27/2020]. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P110020S016
- Qu K, Pan Q, Zhang X, et al. Detection of BRAF V600 mutations in metastatic melanoma: comparison of the Cobas 4800 and Sanger sequencing assays. J Mol Diagn 2013;15:790-5. [Crossref] [PubMed]
- Lopez-Rios F, Angulo B, Gomez B, et al. Comparison of testing methods for the detection of BRAF V600E mutations in malignant melanoma: pre-approval validation study of the companion diagnostic test for vemurafenib. PLoS One 2013;8:e53733. [Crossref] [PubMed]
- Anderson S, Bloom KJ, Vallera DU, et al. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med 2012;136:1385-91. [Crossref] [PubMed]
- Therascreen BRAF V600E RGQ PCR Kit 2020 [updated 7/27/2020]. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P190026
- Moncur JT, Bartley AN, Bridge JA, et al. Performance Comparison of Different Analytic Methods in Proficiency Testing for Mutations in the BRAF, EGFR, and KRAS Genes: A Study of the College of American Pathologists Molecular Oncology Committee. Arch Pathol Lab Med 2019;143:1203-11. [Crossref] [PubMed]
- Surrey LF, Oakley FD, Merker JD, et al. Next-Generation Sequencing (NGS) Methods Show Superior or Equivalent Performance to Non-NGS Methods on BRAF, EGFR, and KRAS Proficiency Testing Samples. Arch Pathol Lab Med 2019;143:980-4. [Crossref] [PubMed]
- Pennell NA, Mutebi A, Zhou ZY, et al. Economic Impact of Next-Generation Sequencing Versus Single-Gene Testing to Detect Genomic Alterations in Metastatic Non–Small-Cell Lung Cancer Using a Decision Analytic Model. JCO Precision Oncology 2019;3:1-9. [Crossref]
- Nucleic Acid Based Tests 2020 [updated 7/7/2020]. Available online: https://www.fda.gov/medical-devices/vitro-diagnostics/nucleic-acid-based-tests
- Pearlstein MV, Zedek DC, Ollila DW, et al. Validation of the VE1 immunostain for the BRAF V600E mutation in melanoma. J Cutan Pathol 2014;41:724-32. [Crossref] [PubMed]
- Schirosi L, Strippoli S, Gaudio F, et al. Is immunohistochemistry of BRAF V600E useful as a screening tool and during progression disease of melanoma patients? BMC Cancer 2016;16:905. [Crossref] [PubMed]
- Fisher KE, Cohen C, Siddiqui MT, et al. Accurate detection of BRAF p.V600E mutations in challenging melanoma specimens requires stringent immunohistochemistry scoring criteria or sensitive molecular assays. Hum Pathol 2014;45:2281-93. [Crossref] [PubMed]
- Rapisuwon S, Busam KJ, Parks K, Chapman PB, Lee E, Atkins MB. Discordance Between Cobas BRAF V600 Testing and VE1 Immunohistochemistry in a Melanoma Patient With Bone Marrow Metastases. Am J Dermatopathol 2016;38:687-9. [Crossref] [PubMed]
- Bourhis A, Le Flahec G, Uguen A. Decalcification can cause the failure of BRAF molecular analyses and anti-BRAFV600E VE1 immunohistochemistry. Pathol Int 2019;69:219-23. [Crossref] [PubMed]
- Zimmermann AK, Camenisch U, Rechsteiner MP, et al. Value of immunohistochemistry in the detection of BRAF(V600E) mutations in fine-needle aspiration biopsies of papillary thyroid carcinoma. Cancer Cytopathol 2014;122:48-58. [Crossref] [PubMed]
- Wobker SE, Kim LT, Hackman TG, et al. Use of BRAF v600e immunocytochemistry on FNA direct smears of papillary thyroid carcinoma. Cancer Cytopathol 2015;123:531-9. [Crossref] [PubMed]
- Straccia P, Brunelli C, Rossi ED, et al. The immunocytochemical expression of VE-1 (BRAF V600E-related) antibody identifies the aggressive variants of papillary thyroid carcinoma on liquid-based cytology. Cytopathology 2019;30:460-7. [Crossref] [PubMed]
- Mesteri I, Bayer G, Meyer J, et al. Improved molecular classification of serrated lesions of the colon by immunohistochemical detection of BRAF V600E. Mod Pathol 2014;27:135-44. [Crossref] [PubMed]
- Kwon JH, Jeong BK, Yoon YS, et al. Utility of BRAF VE1 Immunohistochemistry as a Screening Tool for Colorectal Cancer Harboring BRAF V600E Mutation. J Pathol Transl Med 2018;52:157-63. [Crossref] [PubMed]
- Adackapara CA, Sholl LM, Barletta JA, et al. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology 2013;63:187-93. [Crossref] [PubMed]
- Panarelli NC, Weidner AS, Yantiss RK, et al. A cautionary note on the immunohistochemical detection of BRAF v600e mutations in serrated lesions of the colon. Mod Pathol 2015;28:740-1. [Crossref] [PubMed]
- Estrella JS, Tetzlaff MT, Bassett RL Jr, et al. Assessment of BRAF V600E Status in Colorectal Carcinoma: Tissue-Specific Discordances between Immunohistochemistry and Sequencing. Mol Cancer Ther 2015;14:2887-95. [Crossref] [PubMed]
- Bledsoe JR, Kamionek M, Mino-Kenudson M. BRAF V600E immunohistochemistry is reliable in primary and metastatic colorectal carcinoma regardless of treatment status and shows high intratumoral homogeneity. Am J Surg Pathol 2014;38:1418-28. [Crossref] [PubMed]
- Ilie M, Long E, Hofman V, et al. Diagnostic value of immunohistochemistry for the detection of the BRAFV600E mutation in primary lung adenocarcinoma Caucasian patients. Ann Oncol 2013;24:742-8. [Crossref] [PubMed]
- Sasaki H, Shimizu S, Tani Y, et al. Usefulness of immunohistochemistry for the detection of the BRAF V600E mutation in Japanese lung adenocarcinoma. Lung Cancer 2013;82:51-4. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Mol Diagn 2018;20:129-59. [Crossref] [PubMed]
- Gow CH, Hsieh MS, Lin YT, et al. Validation of Immunohistochemistry for the Detection of BRAF V600E-Mutated Lung Adenocarcinomas. Cancers (Basel) 2019;11:866. [Crossref] [PubMed]
- Hofman V, Benzaquen J, Heeke S, et al. Real-world assessment of the BRAF status in non-squamous cell lung carcinoma using VE1 immunohistochemistry: A single laboratory experience (LPCE, Nice, France). Lung Cancer 2020;145:58-62. [Crossref] [PubMed]
Cite this article as: Sholl LM. A narrative review of BRAF alterations in human tumors: diagnostic and predictive implications. Precis Cancer Med 2020;3:26.

