
非小细胞肺癌的病理学:不断变化的情况
引言
肺癌是全世界癌症死亡的最主要原因[1]。迄今为止,对于非小细胞肺癌(NSCLC)患者的靶向治疗策略有了明显的改善[2-8]。为此,美国病理学家学院(CAP)、国际肺癌研究协会(IASLC)和分子病理学协会(AMP)、国家综合癌症网络(NCCN)和美国临床肿瘤学会(ASCO)指南,定义了晚期NSCLC患者必须检测的一些基因,包括表皮生长因子受体(EGFR)、无性淋巴瘤激酶(ALK)、ROS原癌基因1受体酪氨酸激酶(ROS1)和V-Raf鼠肉瘤病毒癌基因同源物B(BRAF),以便用酪氨酸激酶抑制剂(TKI)进行治疗[9-11]。在这种不断发展的情况下,分子预测病理学在NSCLC患者的管理中起着关键作用[12]。对晚期NSCLC患者的一个重要限制就是可用于形态学和分子学目的的组织标本数量少[13]。特别是在这些患者中,有很高比例的患者只有小的组织活检和细胞学样本可以满足形态学和分子学的要求,而在30%的病例中没有组织标本[14-17]。为了克服这一限制,一种有效的方法是可以采用液体来源的样本来评估这些患者的分子状态,即所谓的“液体活检”[18-20]。通过不同的分析方法,可以从血样以及其他液体标本中评估不同的相关分子生物学分析物的状态,迄今为止,唯一被批准用于NSCLC患者的TKI管理的标志物是以从血浆中提取的循环肿瘤DNA(ctDNA)为代表,用来评估EGFR的状态[13,18]。特别是对于没有组织的患者(肿瘤组织不足或无法获得组织标本),建议对从血浆中提取的ctDNA进行分析,以克服患者不适和活检风险相关的问题[13,18,21-26]。这种方法的一个严重局限是血液中的ctDNA含量较低(<0.5%的总细胞游离DNA),这可能与假阴性结果有关[13,14]。值得注意的是,我们通过二代测序(NGS)方法检测一个小型基因芯片[SiRe®,Genedin(a spin-off of the Department of Public Health, UnⅣersity of Naples “Federico Ⅱ”), Rome, Italy]证实ctDNA和组织结果之间的高度一致性,该芯片涵盖6个基因(EGFR、KRAS、NRAS、BRAF、PDGFRα和c-kit)的568个临床相关突变位点[27-29]。然而,血液样本中的一些分析物(如循环肿瘤细胞、外泌体、血小板RNA和循环肿瘤RNA),在NSCLC治疗决策中显示出重要作用[30-33]。除血液样本外,其他液体可被视为“液体活检”,可能为分子病理学家和临床医生提供NSCLC患者基因评估的相关信息,以指导TKIs的应用。
在本综述中,我们重点关注液体活检方法的先进性,特别是关注这些不同于血液样本的其他来源的液体样本。
尿液
肾小球过滤是血浆的自然“离心过程”,可使尺寸较小的DNA片段,即尿液中存在肿瘤DNA通过[20,34,35]。收集尿液样本与血液样本相比,无创是一个主要优点[20]。而主要的缺点是DNA和RNA水解酶的活性较高,可加快两种分析物的降解[36,37]。Reckamp等人首先在TIGER-X试验中采用了尿样来鉴定EGFR突变,这是一项针对晚期NSCLC患者的第三代TKI(罗西替尼)的1/2期临床研究[38]。通过使用突变富集PCR结合NGS检测,对n=60的尿液样本与匹配的组织标本进行检测,作者显示,当考虑所有尿量时,EGFR第20号外显子p.T790M突变、第21号外显子p.L858R突变和第19号外显子缺失的敏感性分别为72%、75%和67%;当收集推荐尿量为90~100 mL时,敏感性分别为93%、80%和83%;特异性为96%、100%和94%[38]。有趣的是,作者强调了尿液和血浆与组织的互补作用。特别是作者在12个以前在组织样本中检测不到或结果不充分的病例中发现EGFR第20号外显子p.T790M突变(n=10在尿液和血浆样本中,n=1仅在血浆中,n=1仅在尿液中)。其中,n=9显示在服用罗西替尼21天后,EGFR外显子20 p.T790M突变的尿液浓度明显下降[38]。Li等人通过微滴式数字PCR(ddPCR)分析了NSCLC患者不同阶段的尿液、血浆和组织样本[39]。总的来说,作者报告了尿液和组织之间的高度一致性,特别是在晚期(42%的I/Ⅱ期与93%的Ⅲ/Ⅳ期),以及尿液和血浆之间的一致性(75%的Ⅰ/Ⅱ期与100%的Ⅲ/Ⅳ期)[39]。Hu等人用ddPCR比较了213名接受手术的NSCLC患者的尿液样本和组织标本(Ⅰ~Ⅲ期)以及已知的EGFR突变(n=111EGFR第21号外显子p.L858R突变和n=102EGFR第19号外显子缺失)[40]。总的来说,只有n=130的尿样显示EGFR阳性结果(61%;65.8%为EGFR第21号外显子p.L858R突变,55.9%为EGFR第19号外显子缺失)[40]。有趣的是,作者证明在治疗后的患者尿液中都出现了EGFR突变减少的现象[40]。Zhang等人对晚期NSCLC患者的n=160份尿液样本进行了ddPCR,并有相匹配的组织标本[41]。作者显示对EGFR第21号外显子p.L858R突变、EGFR第19号外显子缺失和EGFR第20号外显子p.T790M突变的一致性分别为79%、81%和100%[41]。考虑到所有的EGFR突变,阳性预测值(PPV)和阴性预测值(NPV)分别为100%和53.6%[41]。Wu等人用NGS方法分析了50个晚期(ⅢB/Ⅳ)患者的尿液样本[42]。作者显示组织和尿液之间的一致率为70%[42]。Wang等人用ddPRC方法检测了200个晚期NSCLC患者的尿液和组织样本[43]。作者将注意力集中在KRAS突变上,以达到预后的目的[43]。总的来说,78%(109/140)的病例在两个样本上都显示出KRAS突变;尿液样本中没有出现假阳性结果(两个样本都是阴性的病例=60)[43]。作者还强调,尿液中KRAS突变DNA的鉴定和浓度是预示着更坏的结果[43]。同样,Xie等人分析了n=150病例(n=100个KRAS突变型病例和n=50个KRAS野生型病例),研究尿液分析对预后的潜在作用[44]。以组织结果作为金标准,KRAS阳性组的总体一致性为77%;而野生型组的一致性为92%[44]。作者论述了当执行连续尿液分析时发现越来越多的尿液中KRAS阳性突变的病例(在组织阳性和尿液阴性之前)(93%)[44]。表1总结了尿液样本的结果。
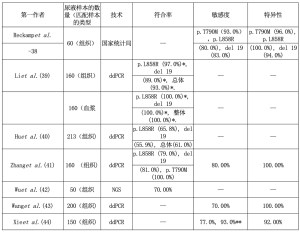
Full table
唾液
唾液中含有多种不同的蛋白质、核酸、电解质和来自不同器官的激素[45]。因此,唾液可以作为生物标志物评估的来源,正如Streckfus等人所显示的那样,他们在乳腺癌患者的唾液标本中检测到c-erbB-2[46]。在Wu等人的经验中,除了尿液样本外,作者还分析了50个唾液样本[42]。有趣的是,作者显示组织和唾液之间的吻合率为74%[42]。Pu等人使用电场诱导释放和测量(EFIRM)技术分析了17份唾液样本,这些样本是在NSCLC患者手术前后收集的[47]。作为黄金标准,作者采用了美国食品和药物管理局(FDA)批准的Cobas检测法对匹配的组织样本进行检测[47]。值得注意的是,所有病例都有19号外显子缺失或EGFR第21号外显子p.L858R突变,所有野生型病例都在唾液样本中得到证实[47]。只有n=1个EGFR第18号外显子p.G719X突变由于没有特异性探针,在唾液中没有被发现[47]。作者证明只有n=1个血浆样品的假阳性结果(EGFR第21号外显子p.L858R突变)[47]。Hubers等对10个EGFR组织突变的病例和20个没有EGFR突变的病例[10个肺癌患者EGFR野生型病例和10个慢性阻塞性肺病(COPD)患者]进行了至少4种不同的测试[循环PCR、低变性温度共同扩增-PCR(COLD-PCR)、Pangaea Biotech SL技术(PST)和高分辨率熔解(HRM)][48]。在唾液样本中没有评估到假阳性结果;此外,50%的敏感性是较高的证据[48]。表2中总结了唾液样本的结果。
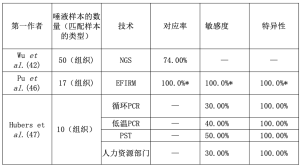
Full table
脑脊液(CSF)
NSCLC患者脑膜转移(LMs)的发生率约为3%至5%,在EGFR突变的患者中发生率更高[49,50]。虽然腰椎穿刺是一个侵入性的过程,但CSF是一个有用的工具,可以获得脑转移的NSCLC患者的细胞游离DNA(cfDNA)[20,51]。Li等人在EGFR突变的NSCLC患者LM中对n=26个CSF样本进行了NGS方法检测[52]。在所有的案例中,从CSF标本中提取的cfDNA中的突变都被正确发现了[52]。此外,在匹配的CSF和血浆样本的沉淀物中也发现了突变[52]。有趣的是,CSF cfDNA中EGFR第20号外显子p.T790M图案变的检测率(7/23,30.4%)高于血浆(5/23,21.7%)[52]。在另一个试验中,同组的Zheng等人分析了n=11份ALK重排阳性患者的CSF和血浆样本[53]。总的来说,CSF样本的一致率(9/11,81.8%)高于血浆(5/11,45.5%)[53]。Ying等人对72个匹配的CSF和血浆样本进行了基于捕获的定向测序[54]。在考虑任何突变(81.5% vs 62.5%)或EGFR突变(58.3% vs 44.4%)时,作者显示CSF的突变检测率高于血浆[54]。在对EGFR突变病例的详细分析中,作者证明在51.4%的CSF样本和38.9%的血浆样本中发现了EGFR激活性突变[54]。Zhao等人重点关注了7名EGFR TKI治疗(吉非替尼)失败的NSCLC患者[55]。特别是与匹配的血浆样本不同,作者显示,由于吉非替尼对血脑屏障的渗透率低,所有CSF样本与血浆相比,其中的EGFR突变克隆都持续地存在(2/7,28.6%)[55]。Kawahara等人证明了检测EGFR突变的高灵敏度、特异性和准确性(分别为87.5%、100%、91.7%),包括EGFR第20外显子20 p.T790M突变,通过使用cobas®EGFR突变测试v2[56]。Yang等人对n=30个肺腺癌脑转移的病例进行了扩增难治性突变系统(ARMS)-PCR检测[57]。作者报告组织和CSF样本之间的PPV、NPV、敏感性和特异性分别为75%、75%、67%和82%;EGFR突变病例的一致率为75%[57]。同样地,Rong等人对5名有EGFR致敏突变的NSCLC患者的CSF采用了ARMS-PCR[58]。只有3例(60%)确认了突变[58]。表3总结了CSF样本的结果。
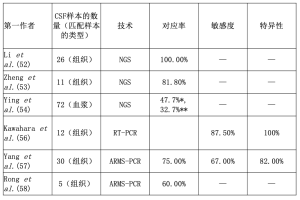
Full table
胸腔积液(PE)
恶性胸腔积液(MPEs)经常出现在晚期NSCLC患者中[59]。虽然胸腔穿刺是一个侵入性的过程,但对于诊断、治疗和分子学的目的来说是必要的[60-62]。Kimura等人首先报道了通过分析从PE中提取的DNA来检测致敏突变的可能性,以及与TKI治疗反应(本文为部分反应)的相关性[63]。Liu等人通过ARMS-PCR对n=41个匹配的转移性胸膜肿瘤组织(MPTTs)、MPE上清液和MPE细胞块(CBs)进行了EGFR分析[64]。总的来说,MPTT和MPE上清液的结果显示,MPE上清液的EGFR突变检测灵敏度和特异性分别为84.2%和90.9%[64]。当考虑2个MPE样本时,观察到更高的灵敏度(94.7%),但特异性较低(81.8%)[64]。Lin等人通过使用HRM分析了22个匹配的MPE上清液、MPE细胞颗粒和组织活检样本[65]。以组织活检结果作为金标准,在MPE上清液样本中没有发现假阳性结果(特异性100%),而只有n=1个假阴性结果被证明(敏感性92.3%)[65]。在MPE细胞颗粒样品中,有较多的假阴性结果(4/22,18.2%)被证明[65]。同样,Kawahara等人通过使用TaqMan突变检测法或基于荧光共振能量转移的优先同源双链形成检测法(F-PHFA)分析了74个(n=29个EGFR野生型和n=45个EGFR突变病例)匹配的MPE上清液、MPE细胞颗粒和组织活检样品[66]。作者报告对MPE上清液的敏感性和特异性分别为44.4%和100%[66]。Lee等人通过使用肽核酸(PNA)介导的PCR夹子对两组肺腺癌(ADC)患者进行EGFR分析[67]。特别是作者分析了从PE中提取的cfDNA,这些cfDNA来自先前基因分型的酪氨酸激酶抑制(TKI-naïve)的患者(n=32;n=19 EGFR突变患者和n=13野生型病例)和TKI-获得性耐药患者(n=18)[67]。在第一组中,2个样本之间的一致性为88%;特别是在从PE中提取的cfDNA中,19个EGFR突变中的2个被遗漏,而在野生型组中发现了2个额外的突变[67]。在第二组中,从PE提取的cfDNA中只有n=1个EGFR突变被遗漏。值得注意的是,在11个(61%)病例中,除了最初的EGFR致敏突变外,还发现了一个额外的EGFR第20号外显子p.T790M突变[67]。总的来说,考虑到EGFR致敏性突变,第二组的一致率为94%[67]。表4总结了PE样本的结果。
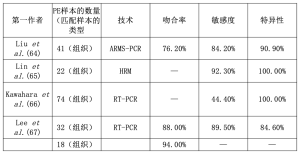
Full table
未来展望和结论
除了“体液活检”外,另一个用于分子目的的cfDNA来源是细胞学制备后的上清液。这种被忽视的材料通常被丢弃在细胞学实验室里。然而,一些研究表明,从细胞学制备的上清液中提取的cfDNA适合于NGS分析[68-74]。
总之,所谓的“液体活检”代表了一种有效的材料,不仅可以替代基于组织的测试,而且可以更好地确定NSCLC患者的生物标志物的分子状态,以确定最佳的治疗选择[75,76]。
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors Alfredo Addeo and Giuseppe Banna for the series “Non-Small Cell Lung Cancer (NSCLC)” published in Precision Cancer Medicine. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.05.04). The series “Non-Small Cell Lung Cancer (NSCLC)” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin 2012;62:10-29. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. J Clin Oncol 2018;36:911-9. [Crossref] [PubMed]
- Ettinger DS, Aisner DL, Wood DE, et al. NCCN Guidelines Insights: Non-Small Cell Lung Cancer, Version 5.2018. J Natl Compr Canc Netw 2018;16:807-821. [Crossref] [PubMed]
- Walk EE. The role of pathologists in the era of personalized medicine. Arch Pathol Lab Med 2009;133:605-10. [PubMed]
- Pisapia P, Malapelle U, Troncone G. Liquid Biopsy and Lung Cancer. Acta Cytol 2018; [Epub ahead of print]. [Crossref] [PubMed]
- Malapelle U, Pisapia P, Rocco D, et al. Next generation sequencing techniques in liquid biopsy: focus on non-small cell lung cancer patients. Transl Lung Cancer Res 2016;5:505-10. [Crossref] [PubMed]
- Ofiara LM, Navasakulpong A, Ezer N, et al. The importance of a satisfactory biopsy for the diagnosis of lung cancer in the era of personalized treatment. Curr Oncol 2012;19:S16-23. [Crossref] [PubMed]
- Wang S, Yu B, Ng CC, et al. The suitability of small biopsy and cytology specimens for EGFR and other mutation testing in non-small cell lung cancer. Transl Lung Cancer Res 2015;4:119-25. [PubMed]
- Malapelle U, Bellevicine C, De Luca C, et al. EGFR mutations detected on cytology samples by a centralized laboratory reliably predict response to gefitinib in non-small cell lung carcinoma patients. Cancer Cytopathol 2013;121:552-60. [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Sacher AG, Komatsubara KM, Oxnard GR. Application of Plasma Genotyping Technologies in Non-Small Cell Lung Cancer: A Practical Review. J Thorac Oncol 2017;12:1344-56. [Crossref] [PubMed]
- Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: a prospective multicenter study in a real-world setting (GFPC study 12-01). Lung Cancer 2014;86:170-3. [Crossref] [PubMed]
- Heerink WJ, de Bock GH, de Jonge GJ, et al. Complication rates of CT-guided trans thoracic lung biopsy: meta-analysis. Eur Radiol 2017;27:138-48. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Cai W, Lin D, Wu C, et al. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Co altered Lung Adenocarcinoma. J Clin Oncol 2015;33:3701-9. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution ofNon-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Malapelle U, Mayo de-Las-Casas C, Rocco D, et al. Development of a gene panel for next-generation sequencing of clinically relevant mutations in cell-free DNA from cancer patients. Br J Cancer 2017;116:802-10. [Crossref] [PubMed]
- Pisapia P, Pepe F, Smeraglio R, et al. Cell free DNA analysis by SiRe(®) next generation sequencing panel in non small cell lung cancer patients: focus on basal setting. J Thorac Dis 2017;9:S1383-90. [Crossref] [PubMed]
- Pisapia P, Rocco D, Pepe F, et al. EGFR exon 19 deletion switch and development of p.L792Q mutation as a new resistance mechanism to osimertinib: a case report and literature review. Transl Cancer Res 2019;8:S64-9. [Crossref]
- Joosse SA, Pantel K. Tumor-Educated Platelets as Liquid Biopsy in Cancer Patients. Cancer Cell 2015;28:552-4. [Crossref] [PubMed]
- Raez LE, Manca P, Rolfo C, et al. ROS-1 Rearrangements in Circulating Tumor Cells. J Thorac Oncol 2018;13:e71-2. [Crossref] [PubMed]
- Reclusa P, Taverna S, Pucci M, et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J Thorac Dis 2017;9:S1373-82. [Crossref] [PubMed]
- Reclusa P, Laes JF, Malapelle U, et al. EML4-ALK translocation identification in RNA exosomal cargo (ExoALK) in NSCLC patients: a novel role for liquid biopsy. Transl Cancer Res 2019;8:S76-8. [Crossref]
- Su YH, Wang M, Brenner DE, et al. Human urine contains small, 150 to 250 nucleotide-sized, soluble DNA derived from the circulation and may be useful in the detection of colorectal cancer. J Mol Diagn 2004;6:101-7. [Crossref] [PubMed]
- Su YH, Wang M, Aiamkitsumrit B, et al. Detection of a K-ras mutation in urine of patients with colorectal cancer. Cancer Biomark 2005;1:177-82. [Crossref] [PubMed]
- Nadano D, Yasuda T, Kishi K. Measurement of deoxyribonuclease I activity inhuman tissues and body fluids by a single radial enzyme-diffusion method. Clin Chem 1993;39:448-52. [PubMed]
- Mall C, Rocke DM, Durbin-Johnson B, et al. Stability of miRNA in human urine supports its biomarker potential. Biomark Med 2013;7:623-31. [Crossref] [PubMed]
- Reckamp KL, Melnikova VO, Karlovich C, et al. A Highly Sensitive and Quantitative Test Platform for Detection of NSCLC EGFR Mutations in Urine and Plasma. J Thorac Oncol 2016;11:1690-700. [Crossref] [PubMed]
- Li F, Huang J, Ji D, et al. Utility of urinary circulating tumor DNA for EGFR mutation detection in different stages of non-small cell lung cancer patients. Clin Transl Oncol 2017;19:1283-91. [Crossref] [PubMed]
- Hu T, Shen H, Huang H, et al. Urinary circulating DNA profiling in non-small cell lung cancer patients following treatment shows prognostic potential. J Thorac Dis 2018;10:4137-46. [Crossref] [PubMed]
- Zhang H, He B, Cui J, et al. Comparison of circulating DNA from plasma and urine for EGFR mutations in NSCLC patients. Cancer Biomark 2018;23:427-36. [Crossref] [PubMed]
- Wu Z, Yang Z, Li CS, et al. Differences in the genomic profiles of cell-free DNA between plasma, sputum, urine, and tumor tissue in advanced NSCLC. Cancer Med 2019;8:910-9. [Crossref] [PubMed]
- Wang X, Meng Q, Wang C, et al. Investigation of transrenal KRAS mutation in late stage NSCLC patients correlates to disease progression. Biomarkers 2017;22:654-60. [PubMed]
- Xie F, Li P, Gong J, et al. Urinary cell-free DNA as a prognostic marker for KRAS-positive advanced-stage NSCLC. Clin Transl Oncol 2018;20:591-8. [Crossref] [PubMed]
- Gao K, Zhou H, Zhang L, et al. Systemic disease-induced salivary biomarker profiles in mouse models of melanoma and non-small cell lung cancer. PLoS One 2009;4:e5875. [Crossref] [PubMed]
- Streckfus C, Bigler L, Dellinger T, et al. The presence of soluble c-erbB-2 in saliva and serum among women with breast carcinoma: a preliminary study. Clin Cancer Res 2000;6:2363-70. [PubMed]
- Pu D, Liang H, Wei F, et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac Cancer 2016;7:428-36. [Crossref] [PubMed]
- Hubers AJ, Heideman DA, Yatabe Y, et al. EGFR mutation analysis in sputum of lung cancer patients: a multi technique study. Lung Cancer 2013;82:38-43. [Crossref] [PubMed]
- Remon J, Le Rhun E, Besse B. Leptomeningeal carcinomatosis in non-small cell lung cancer patients: A continuing challenge in the personalized treatment era. Cancer Treat Rev 2017;53:128-37. [Crossref] [PubMed]
- Li YS, Jiang BY, Yang JJ, et al. Leptomeningeal Metastases in Patients with NSCLC with EGFR Mutations. J Thorac Oncol 2016;11:1962-9. [Crossref] [PubMed]
- Pan W, Gu W, Nagpal S, et al. Brain tumor mutations detected in cerebral spinal fluid. Clin Chem 2015;61:514-22. [Crossref] [PubMed]
- Li YS, Jiang BY, Yang JJ, et al. Unique genetic profiles from cerebrospinal fluid cell-free DNA in leptomeningeal metastases of EGFR-mutant non-small-cell lung cancer: a new medium of liquid biopsy. Ann Oncol. 2018;29:945-52. [Crossref] [PubMed]
- Zheng MM, Li YS, Jiang BY, et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J Thorac Oncol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Ying S, Ke H, Ding Y, et al. Unique genomic profiles obtained from cerebrospinal fluid cell-free DNA of non-small cell lung cancer patients with leptomeningeal metastases. Cancer Biol Ther 2019;20:562-70. [Crossref] [PubMed]
- Zhao J, Ye X, Xu Y, et al. EGFR mutation status of paired cerebrospinal fluid and plasma samples in EGFR mutant non-small cell lung cancer with leptomeningeal metastases. Cancer Chemother Pharmacol 2016;78:1305-10. [Crossref] [PubMed]
- Kawahara A, Abe H, Murata K, et al. Screening system for epidermal growth factor receptor mutation detection in cytology cell-free DNA of cerebrospinal fluid based on assured sample quality. Cytopathology 2019;30:144-9. [Crossref] [PubMed]
- Yang H, Cai L, Zhang Y, et al. Sensitive detection of EGFR mutations in cerebrospinal fluid from lung adenocarcinoma patients with brain metastases. J Mol Diagn 2014;16:558-63. [Crossref] [PubMed]
- Rong J, Chunhua M, Yuan L, et al. Detected EGFR mutation in cerebrospinal fluid of lung adenocarcinoma patients with meningeal metastasis. Open Med (Wars) 2016;11:93-6. [Crossref] [PubMed]
- Antony VB, Loddenkemper R, Astoul P, et al. Management of malignant pleural effusions. Eur Respir J 2001;18:402-19. [Crossref] [PubMed]
- Dai Y, Morishita Y, Mase K, et al. Application of the p53 and K-ras gene mutation patterns for cytologic diagnosis of recurrent lung carcinomas. Cancer 2000;90:258-63. [Crossref] [PubMed]
- Huang MJ, Lim KH, Tzen CY, et al. EGFR mutations in malignant pleural effusion of non-small cell lung cancer: a case report. Lung Cancer 2005;49:413-5. [Crossref] [PubMed]
- Soh J, Toyooka S, Aoe K, et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer 2006;119:2353-8. [Crossref] [PubMed]
- Kimura H, Fujiwara Y, Sone T, et al. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer 2006;95:1390-5. [Crossref] [PubMed]
- Liu D, Lu Y, Hu Z, et al. Malignant pleural effusion supernatants are substitutes for metastatic pleural tumor tissues in EGFR mutation test in patients with advanced lung adenocarcinoma. PLoS One 2014;9:e89946. [Crossref] [PubMed]
- Lin J, Gu Y, Du R, et al. Detection of EGFR mutation in supernatant, cell pellets of pleural effusion and tumor tissues from non-small cell lung cancer patients by high resolution melting analysis and sequencing. Int J Clin Exp Pathol 2014;7:8813-22. [PubMed]
- Kawahara A, Fukumitsu C, Azuma K, et al. A Combined test using both cell sediment and supernatant cell-free DNA in pleural effusion shows increased sensitivity in detecting activating EGFR mutation in lung cancer patients. Cytopathology 2018;29:150-5. [Crossref] [PubMed]
- Lee JS, Hur JY, Kim IA, et al. Liquid biopsy using the supernatant of a pleural effusion for EGFR genotyping in pulmonary adenocarcinoma patients: a comparison between cell-free DNA and extracellular vesicle-derived DNA. BMC Cancer 2018;18:1236. [Crossref] [PubMed]
- Tian SK, Killian JK, Rekhtman N, et al. Optimizing Workflows and Processing of Cytologic Samples for Comprehensive Analysis by Next-Generation Sequencing: Memorial Sloan Kettering Cancer Center Experience. Arch Pathol Lab Med 2016; [Epub ahead of print]. [Crossref] [PubMed]
- Roy-Chowdhuri S. Molecular testing of residual cytology samples: Rethink, reclaim, repurpose. Cancer Cytopathol 2019;127:15-7. [Crossref] [PubMed]
- Rekhtman N, Roy-Chowdhuri S. Cytology Specimens: A Goldmine for Molecular Testing. Arch Pathol Lab Med 2016;140:1189-90. [Crossref] [PubMed]
- Roy-Chowdhuri S, Mehrotra M, Bolivar AM, et al. Salvaging the supernatant: next generation cytopathology for solid tumor mutation profiling. Mod Pathol 2018;31:1036-45. [Crossref] [PubMed]
- Guibert N, Tsukada H, Hwang DH, et al. Liquid biopsy of fine-needle aspiration supernatant for lung cancer genotyping. Lung Cancer 2018;122:72-5. [Crossref] [PubMed]
- Hannigan B, Ye W, Mehrotra M, et al. Liquid Biopsy Assay for Lung Carcinoma Using Centrifuged Supernatants from Fine Needle Aspiration Specimens. Ann Oncol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Ye W, Hannigan B, Zalles S, et al. Centrifuged supernatants from FNA provide a liquid biopsy option for clinical next-generation sequencing of thyroid nodules. Cancer Cytopathol 2019;127:146-60. [Crossref] [PubMed]
- Liquid Biopsy Holds Its Own in NSCLC. Cancer Discov 2019; [Epub ahead of print].
- Leighl NB, Page RD, Raymond VM, et al. Clinical Utility of Comprehensive Cell-Free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-Small Cell Lung Cancer. Clin Cancer Res 2019; [Epub ahead of print]. [Crossref] [PubMed]

张冲
中共党员,博士,病理学在站博士后。中国医学装备人工智能联盟病理委员会第一届委员,重庆市免疫学会代谢免疫专委会第一届常务委员,《转化医学电子杂志》编辑委员会第二届青年编委。2018-2019年涪陵区卫健委“优秀共产党员”。主持2020年中国博士后科学基金面上资助1项,2020年涪陵区新冠疫情应急防治科技攻关专项项目1项,2019年度重庆市基础研究与前沿探索专项博士后科学基金项目1项,2018年重庆市博士后特别资助三等资助项目1项,2017年重庆市卫计委科研项目1项,参译《食管癌》(ISBN 978-7-5487-2238-02018)一书,发表论文8篇(SCI 4篇)。入选“第58批中国博士后科技服务团(广西钦州行)”。2019年遵义医科大学青年教师讲课比赛三等奖。(更新时间:2021/7/23)
(本译文仅供学术交流,实际内容请以英文原文为准。)
Cite this article as: Pisapia P, Pepe F, Iaccarino A, Troncone G, Malapelle U. Pathology in non-small cell lung cancer: evolving scenario. Precis Cancer Med 2019;2:17.

