
Long non-coding RNA MALAT1 as metastasis suppressor
Treatment of the metastatic patient is the most unmet clinical need in cancer treatment. While the genomic mutational landscape is fundamental to design precise medicine therapeutic approaches for different primary human tumors, its contribution to the treatment of metastatic dissemination is uncertain. This paves the way for intense research on metastasis by exploring alternative molecular landscapes, including the non-coding RNA (NCR) layer. Kim et al. have recently shown that lncRNA-MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), an abundant lncRNA expressed in several tumors and involved in tissue repair, operates as metastasis suppressor in breast cancer (BC) (1). BC is one of the most frequent tumors worldwide whose lethality is largely due to eruption of aggressive, chemoresistant metastases (mBC), occurring rapidly in a large fraction of patients (37%) bearing triple negative BC (TNBC) (2). Using in vivo models (transgenic, xenograft and syngeneic mouse model), Ma’s group showed that overexpression of lncRNA-MALAT1 suppressed BC metastasis (1). Mechanistically, metastasis suppressor activity of lncRNA-MALAT1 intersects with the activity of the YAP/TAZ transcriptional coactivators (Figure 1) (1). These factors are known to serve as mediators of the Hippo cascade, originally identified as a pathway strongly involved in the control of organ size and development. More recently its pivotal role in the cancer insurgence and metastatic dissemination is rapidly emerging (3,4). Seminal work from Piccolo’s group has proved that YAP/TAZ is critical mechanosensors whose aberrant activation contributes to BC metastatization (Figure 1) (5). Notably, Ma’s group elegantly showed, at least under specific experimental conditions, that lcnRNA-MALAT1 interacts with the members of TEAD protein family (1). These are the DNA binding platforms of YAP/TAZ (4,5). The binding of lncRNA-MALAT1 to TEAD prevented its binding to YAP thereby strongly impairing the aberrant transcription of pro-metastatic gene programs (Figure 1) (1,5). Interestingly, TEAD binding sites are present in different regions on lncRNA-MALAT1 suggesting a tight interaction that may underline tight functional control. The interaction with TEAD was rather specific, while no binding was evidenced for YAP itself. Poorly explored, however, is the biochemical and functional associations between MALAT1 and the YAP sibling factor TAZ (5). Ectopic expression of lncRNA-MALAT1 displaced either TEAD or YAP recruitment from canonical target promoters, thereby impairing its pro-metastatic transcriptional potential (Figure 1).
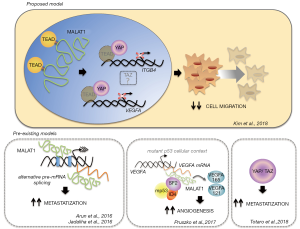
How does the intersection between MALAT1 and TEAD/YAP signaling impact on metastasis? Ma and colleagues focussed on two TEAD target genes that are also two-well characterized metastasis inducers, ITGB4 and VEGFA (Figure 1) (1,6). The latter is a major determinant in tumor neo-angiogenesis and is heavily subjected to alternative splice selection (6). Two families of VEGFA isoforms are indeed generated by alternative splicing of the gene distal exon. Intriguingly, proximal splice selection generates VEGF pro-angiogenic isoforms while those originating by distal splice selection are anti-angiogenic. Pruszko et al. have shown that aberrant recruitment of lncRNA-MALAT1 on VEGF pre-mRNA dictated VEGF isoform expression (Figure 1) (7). A quaternary ribonucleoprotein complex including lncRNA-MALAT1, the oncogenic splicing factor SRF1, gain of function mutant p53 protein and ID4 was recruited at the VEGF pre-mRNA (Figure 1) (7). Basal-like BC carrying TP53 mutations express VEGF angiogenic signature that associates with elevated ID4 expression (7).
These findings question previous published data largely reporting that lncRNA-MALAT1 is associated to metastasis insurgence and progression (Figure 1) (8-12). Conflicting data are rather frequent when analysing the effects due to aberrant modulation, either down-regulation or up-regulation of a given microRNA and lncRNA. Cell and tumor type specificity are potentially involved in such conflicting set of reports related to NCR factors. Moreover, expression levels, affinity to target site, efficiency of sponge activity, subcellular localization, stability, binding to DNA and other undiscovered properties of NCR factors might represent additional sources of discrepancies.
There is potentially important translational value in the work of Ma and colleagues. BC patients remain at risk of experiencing metastasis for their entire lifetime; although only a fraction of them will relapse, this risk cannot be accurately predicted in individual patients, and thus adjuvant chemotherapy is currently offered to most patients. This raises the need of new prognostic markers of the metastatic disease to avoid overtreatment. NCR factors might be ideal non-invasive biomarkers. Indeed, they circulate in body fluids such as blood, saliva, urines mostly coated in exosomes in which RNA processing occurs and might contribute to the establishment of the metastatic site. Liquid biopsy approaches in which the mutational analysis of cell-free circulating DNA will be combined with the detection of specific NCR factors, such as lncRNA-MALAT1, might be much more accurate in monitoring the metastatic process and the response to therapy.
This body of work leaves unanswered a number of questions. Still unclear is whether lncRNA-MALAT1 expression levels may be positively or negatively correlated with metastatization. As previously mentioned, metastatic dissemination is still poorly known. It is also a very complex process that cannot be fully understood if it is investigated independently from homing, that is, the site of metastatization. BC metastasizes to lung, brain, bone, liver and the molecular mechanisms underlying specific metastatic homing are rather unknown. This might also imply that lung metastasis which is reported from Ma’s does not fully recapitulate the wide range of molecular events underlying the metastatic process caused by an advanced BC. A large body of evidence has shown that microenvironment with its cellular components and released cytokines contributes strongly to determine dissemination and chemoresistance of a primary tumor. Little is known about the crosstalk between metastatic tumor cell and its surrounding microenvironment and how NCR layer may modify all this. Similarly, the intersection of the genomic or epigenomic landscape with its non-coding and coding components to the full deciphering of a metastatic dissemination is only in its infancy. Collectively, it appears mandatory to explore further, to challenge existing models, but it might be too early to call for model rectification.
AcknowledgmentsOther Section
Contribution of AIRC IG 2017 to G Blandino, of Lazio Innova to G Blandino, and of Bilateral Italy-USA to G Blandino, was greatly appreciated.
Funding: None.
FootnoteOther Section
Provenance and Peer Review: This article was commissioned by the editorial office, Precision Cancer Medicine. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/pcm.2019.02.02). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
ReferencesOther Section
- Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 2018;50:1705-15. [Crossref] [PubMed]
- Garrido-Castro AC, Lin NU, Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov 2019;9:176-98. [Crossref] [PubMed]
- Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016;29:783-803. [Crossref] [PubMed]
- Lo Sardo F, Strano S, Blandino G. YAP and TAZ in Lung Cancer: Oncogenic Role and Clinical Targeting. Cancers (Basel) 2018;6:10. [PubMed]
- Totaro A, Panciera T, Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 2018;20:888-99. [Crossref] [PubMed]
- Nowak DG, Amin EM, Rennel ES, et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem 2010;285:5532-40. [Crossref] [PubMed]
- Pruszko M, Milano E, Forcato M, et al. The mutant p53-ID4 complex controls VEGFA isoforms by recruiting lncRNA MALAT1. EMBO Rep 2017;18:1331-51. [Crossref] [PubMed]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22:8031-41. [Crossref] [PubMed]
- Zong X, Tripathi V, Prasanth KV. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol 2011;8:968-77. [Crossref] [PubMed]
- Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med 2013;91:791-801. [Crossref] [PubMed]
- Arun G, Diermeier S, Akerman M, et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016;30:34-51. [Crossref] [PubMed]
- Jadaliha M, Zong X, Malakar P, et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016;7:40418-36. [Crossref] [PubMed]
Cite this article as: Strano S, Donzelli S, Blandino G. Long non-coding RNA MALAT1 as metastasis suppressor. Precis Cancer Med 2019;2:4.

